DNA molecular weight markers
DNA molecular weight markers, commonly referred to as DNA ladders, are essential components in molecular biology experiments. These markers enable precise measurements of nucleic acid fragments in a variety of procedures such as enzymatic digests, PCR amplification, Northern and Southern blotting, and any other application involving gel electrophoresis. Moreover, these molecular weight ladders may be applied not only to nucleic acid size detection but for exploring polyphosphates — inorganic polymeric compounds playing a role in various biological processes [1] Most researchers are currently opting for ready-made commercial molecular weight markers for gel electrophoresis.
DNA ladders
Methods to prepare
Typically, two main methods to create marker sets are used: digesting of custom plasmid by restriction exonucleases and PCR synthesis of DNA fragments. Both methods have several restrictions — for example, restrictase digest of natural DNA produces fragments of irregular length; PCR DNA ladder doesn't fit for detection of small DNA chains (100 – 500 bp). Thus, the effective way to produce appropriate ladders is to combine PCR and partial enzymatic digest.
Sources
Commonly DNA molecules used for preparing marker sets are popular biotechnology objects or constructions such as lambda phage DNA or pBR322 plasmid. In some cases unusual sources can be implicated as a template for DNA ladder. For instance centromeric satellite DNA from beetle Tenebrio molitor may serve as such a template since it contains multiple copies of tandemly repeated fragments with EcoR1 endonuclease site. Partial restriction by this only enzyme produces several types of DNA markers [2]
Commonly DNA molecules used for preparing marker sets are popular biotechnology objects or constructions such as lambda phage DNA or pBR322 plasmid. In some cases unusual sources can be implicated as a template for DNA ladder. For instance centromeric satellite DNA from beetle Tenebrio molitor may serve as such a template since it contains multiple copies of tandemly repeated fragments with EcoR1 endonuclease site. Partial restriction by this only enzyme produces several types of DNA markers [2]
Sizes and ranges
Given the diverse size range of fragments, spanning from tens to thousands base pairs, it is common to utilize two or more sets of molecular weight standards. Typically, ladders with increments of 100 base pairs cover fragment sizes from 100 bp to 1000 bp, while 1 kb plus ladders encompass a range of 500 to several thousand base pairs. Also, long marker DNA ladders with various width spaces in one set, e. g. 250, 500 and 1000 bp (see Sky-High set of Biolabmix) are very useful, when you have a lack of tracks in your gel. Initially the lengths of DNA fragments repeated the natural DNA restriction maps, whereas most modern DNA ladders contain fragments with rounded number size - 100, 200, 300 bp, etc. Nevertheless, in many cases, it is convenient to use sets with a "natural digest", e. g. molecular weight ladder from Biosan pBlueSK/MspI to precise length determination.
Given the diverse size range of fragments, spanning from tens to thousands base pairs, it is common to utilize two or more sets of molecular weight standards. Typically, ladders with increments of 100 base pairs cover fragment sizes from 100 bp to 1000 bp, while 1 kb plus ladders encompass a range of 500 to several thousand base pairs. Also, long marker DNA ladders with various width spaces in one set, e. g. 250, 500 and 1000 bp (see Sky-High set of Biolabmix) are very useful, when you have a lack of tracks in your gel. Initially the lengths of DNA fragments repeated the natural DNA restriction maps, whereas most modern DNA ladders contain fragments with rounded number size - 100, 200, 300 bp, etc. Nevertheless, in many cases, it is convenient to use sets with a "natural digest", e. g. molecular weight ladder from Biosan pBlueSK/MspI to precise length determination.
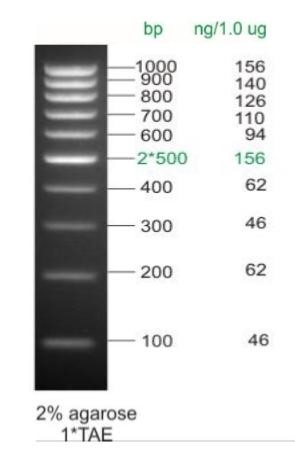
Fig. DNA ladder Step 100 stained with ethidium bromide (Biolabmix)
The central band (or two non-adjacent bands) of the ladder often contains double amount of matter to facilitate its identification within gel.
Marker ladders are suitable both for agarose gel electrophoresis, PAGE (polyacrylamide gel electrophoresis) and pulsed-field gel electrophoresis (PFGE). In the majority of cases, agarose is the preferred option , but it should be noted that the identification of short DNA fragments, e. g. 50 bp DNA ladders will be more accurate in PAGE, whereas long molecules of length above 10000 bp are better resolved in PFGE.
Marker ladders are suitable both for agarose gel electrophoresis, PAGE (polyacrylamide gel electrophoresis) and pulsed-field gel electrophoresis (PFGE). In the majority of cases, agarose is the preferred option , but it should be noted that the identification of short DNA fragments, e. g. 50 bp DNA ladders will be more accurate in PAGE, whereas long molecules of length above 10000 bp are better resolved in PFGE.